30 September 2022: Original Paper
Network Pharmacology-Based Exploration of the Therapeutic Mechanisms of in Renal Ischemia/Reperfusion
Jiajun Dong1BCEF, Mingyang Cao2BCEF, Hui Yu3BCEF, Yang Dong24AD, Conghui Han124AG*DOI: 10.12659/AOT.937469
Ann Transplant 2022; 27:e937469
Abstract
BACKGROUND: Cordyceps cicadae is beneficial in treating renal diseases, especially in inhibiting renal ischemia/reperfusion injury (IRI). The aim of this study was to systematically analyze and predict the potential mechanism of Cordyceps cicadae in renal IRI therapy using network pharmacology.
MATERIAL AND METHODS: Cordycepin, adenosine, and cordycepic acid are the 3 major medicinal ingredients in Cordyceps cicadae. Based on network pharmacology, the 3D structure of the 3 compounds were obtained, and then the common targets between these compounds and renal IRI were analyzed and determined. We used the ingredient-target (I-T), protein–protein interaction (PPI) networks, the enrichment analysis of Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) to find the possible pharmacological mechanism of Cordyceps cicadae in treating renal IRI.
RESULTS: Through target fishing and analysis, the 3 active ingredients of Cordyceps cicadae shared 81 target genes with renal IRI. I-T network showed that adenosine had the highest degree, and 5 genes were associated with the 3 active ingredients. PPI network analysis showed that ALB, GAPDH, CASP3, MAPK1, FN1, and IL-10 play a pivotal role. The enrichment analysis of GO and KEGG showed that Cordyceps cicadae can treat renal IRI through MAPK, cAMP, PPAR, Rap1, and HIF-1 signaling pathways.
CONCLUSIONS: Cordyceps cicadae exerts its therapeutic effect on renal IRI via multiple targets and pathways. Nevertheless, further experimentation is needed to verify this. The method of network pharmacology provides an effective method of determining the comprehensive action mechanism of Traditional Chinese Medicine (TCM).
Keywords: Cordyceps cicadae, Forecasting, Molecular Mechanisms of Pharmacological Action, systems biology, Caspase 3, Cordyceps, Humans, Interleukin-10, Ischemia, Kidney Diseases, Network Pharmacology, peroxisome proliferator-activated receptors, Reperfusion
Background
Ischemia reperfusion injury (IRI) refers to a pathological condition in which tissue or organ damage is aggravated instead of alleviated when the tissue or organ regains perfusion or oxygen supply after ischemia [1]. Clinically, renal IRI occurs most commonly following traumatic shock, sepsis, partial nephrectomy, post-partum hemorrhage, and other major surgical procedures [2]. Surprisingly, the probability of IRI in kidney transplant patients is almost 100% [3]. Although the mechanism by which renal IRI occurs is unclear, it is thought to result from apoptosis, necrosis, oxygen free radicals, inflammation, mitochondrial damage, and ferroptosis [4–6]. One of the key factors is the inflammatory cascade induced by immune cells infiltration [7]. Renal ischemia causes significant tissue hypoxia, which triggers the recruitment of immune cells such as dendritic cells (DC cells), neutrophils, macrophages, and natural killer T (NKT) cells to the injured tissues and the release of pro-inflammatory factors [8]. When blood perfusion is restored later, the inflammatory response is further exacerbated, resulting in progressive aggravation of renal damage [9].
Currently, there is still no satisfactory treatment for IRI. On the one hand, new treatments are effective but have limited clinical applications due to insufficient clinical evidence and serious complications [10,11]. On the other hand, the lack of expertise in renal transplantation pathology has affected the evaluation of the IRI caused by transplanted kidneys to some extent [12], although the application of artificial intelligence is expected to improve such events [13,14]. As a traditional form of treatment, pharmacotherapy has always been the focus of research, but despite this, there are no specific drugs available for treating renal IRI [15,16]. Traditional Chinese Medicine (TCM) treatment has the characteristics of holistic management, multi-targets, multi-pathways, and multi-links, and has become the primary source for the exploration of new drugs by researchers. There are a variety of TCMs that can treat kidney failure, such as
Network pharmacology is a method for network analysis of biological systems based on the theory of systems biology. At present, network pharmacology, as a new and advanced analytical technique, is widely used in pharmacological research, with its superior reliability and efficiency, and is the current research frontier of TCM [24]. Network pharmacology integrates the ideas of systems biology and multidirectional pharmacology to analyze the mechanism of action of drugs by constructing complex drug-ingredient–target-disease networks, enabling pharmacological research to shift from the traditional search for a single target to comprehensive network analysis [25]. The complex chemical composition and multi-target and multi-level pharmacological effects of Chinese herbs are naturally compatible with network pharmacology. In this study, we comprehensively analyzed the mechanism of action of
The purpose of this study was to comprehensively uncover the active ingredients, distinct targets, and accurate molecular mechanism of
Material and Methods
:
With reference to published articles, cordycepin, adenosine, and cordycepic acid were considered likely to be the 3 key active ingredients of Cordyceps cicadae that exert therapeutic effects by targeting diverse proteins. Subsequently, a crucial step was to identify the molecular targets that could disclose the pharmacological mechanisms of Cordyceps cicadae. In this study, using the 3D structural data of the 3 ingredients (Figure 1) obtained in PubChem (PubChem: https://pubchem.ncbi.nlm.nih.gov/), the target prediction was completed based on the PhammerMap database (PhammerMap: http://lilab-ecust.cn/pharmmapper/) and the SwissTargetPrediction database (SwissTargetPrediction: http://www.swisstargetprediction.ch/). Due to the irregular description of the identified candidate targets, the UniProtKB database (UniProtKB: www.uniprot.org/) [26] was used to standardize the candidate targets under the category of “Homo sapiens”. Finally, the unified candidate targets and their gene symbols were obtained from the database.
IDENTIFYING THE TARGETS OF RENAL IR:
The target genes related to renal IR were screened from GeneCards (http://www.genecards.org/). This human gene database is user-friendly, integrative, and searchable. Integrative information in relation to all annotated human diseases, proteins, and genes is available from this database [27]. The database encompasses diverse resources from 125 databases, including NCBI, HGNC, UniProtKB, and ENSEMBL, as well as multitudinous other relevant databases [28]. It is highly reliable in terms of data reliability. Using GeneCards, the information about related targets can be easily retrieved by keyword search, and we identified 1497 renal IR-related targets using the keywords “kidney ischemia reperfusion” and “renal ischemia reperfusion”.
CONSTRUCTING THE INGREDIENT–TARGET (I-T) NETWORK:
To better understand and analyze the molecular mechanism, the visualization software Cytoscape 3.7.2 was used in this study to construct an I-T network. The candidate ingredients targets and renal IR targets were retrieved to obtain the related targets shared with them. The ingredients and the shared targets were then used to map the I-T interaction network using the software. An attribute circle layout algorithm was used when building the network. To arrange each node reasonably and achieve clearer and understandable visualization effect, users can set each node and symbol with appropriate geometric position and set the network topology with personalized graphics and colors through Cytoscape [29]. The importance of every target and ingredient can be estimated by the 2 most critical parameters, degree and betweenness centrality, of the topology. Consequently, the ingredients and targets of Cordyceps cicadae exerting the core effect against renal IR were analyzed and identified.
CONSTRUCTING THE PROTEIN–PROTEIN INTERACTION (PPI) NETWORK:
It is difficult to individually identify specific proteins functions; the proteins mostly constitute macromolecular complexes by interacting with each other in intracellular biochemical processes to complete biological functions. Exploring protein–protein interactions is essential to reveal pharmacodynamic mechanisms, optimize drug efficacy, and reduce adverse effects. To systematically clarify the protein–protein interactions, the relevant targets were retrieved using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; Version 11.0; https://string-db.org/) to construct the PPI network. The STRING database is generally applied to retrieve and construct PPI networks, and its results are derived from experimental research data, literature mining, bioinformatics predictive analysis, and other relevant databases [30]. With STRING’s inherent scoring criteria, the higher the score setting, the higher the reliability of the predicted PPI results. Hence, the lowest score was set at a confidence of 0.4 to guarantee the reliability of the prediction results. The proteins isolated were removed from the network. Finally, the PPI network was the output, and statistical analyses of the PPIs were conducted. The proteins in the network are represented by nodes, while the edges indicate the association among proteins.
GO AND KEGG ENRICHMENT ANALYSIS:
GO analysis is a common approach to describe and annotate the biological processes, cellular components, and molecular functions of genes and gene products [31]. The KEGG is a useful resource that helps to systematically analyze gene functions and related advanced genome functions information [32,33]. In the study, we mainly explored the biological effects of Cordyceps cicadae and realized enrichment analysis by using 3 packages of R software (version “3.6”): (1) “DOSE”, an R package that explores the similarities in diseases and gene functions from a disease perspective by computing semantic similarity involving genes and disease ontology terms [34]; (2) “clusterProfiler”, an R package that mainly functions to compare biological themes and the enrichment analyses of gene clusters [35]; and (3) “pathview”, the R package that can visualize as well as integrate data according to the known pathways [36].
Results
TARGET IDENTIFICATION AND ANALYSIS:
After previous target fishing, it was forecast that 288 targets interacted with the 3 active compounds identified in Cordyceps cicadae. Using keywords retrieval, 1497 genes with relevant to the pathogenesis and progression of renal IR were screened by GeneCards database. By combining the active compounds targets of Cordyceps cicadae with renal IR-related targets, 81 targets shared among them were identified to be potential targets for treating renal IR (Figure 2A), and the details are presented in Supplementary Table 1. From the perspective of network topology, the key targets of the network are regarded as the cores, so the 81 targets determined in this study could be considered as the effective targets for Cordyceps cicadae in the treatment of renal IR.
I-T NETWORK CONSTRUCTION AND ANALYSIS:
To comprehend the relationships between the active components in Cordyceps cicadae and their potential targets common to renal IR, we constructed an I-T network of interactions between ingredients and targets, where every one of the ingredients was connected to its potential targets. The targets, interactions, and active ingredients presented in Figure 2B refer to the 3 active ingredients of Cordyceps cicadae respectively mapped to 81 potential targets. The ingredients and targets are indicated by yellow nodes and green nodes, respectively. And the interactions among the nodes are represented by the edges. The degree of an ingredient node is usually considered to reflect its potential value in drugs. The I-T network analysis showed that adenosine had the highest degree (degree=50), followed by cordycepic acid (degree=36) and cordycepin (degree=13), suggesting the priority of their potential value. Moreover, out of the 81 potential target genes, a total of 8 genes (IL-10, TGM3, MAOA, SMURF2, DPP4, CA2, CA1, and CA9) were linked to 2 ingredients, and 5 genes (TGM2, ERBB4, PTGIS, IGF1R, and SIRT3) were linked to 3 ingredients, indicating that these genes might be crucial in repairing renal IR.
PPI NETWORK CONSTRUCTION AND ANALYSIS:
To better analyze and understand the therapeutic mechanism of Cordyceps cicadae, the PPI network of the targets for the treatment of renal IR was constructed. The confidence level was set above 0.4, and the proteins independent of the network were removed. Then, 75 proteins with 377 relationships were obtained by PPI network analysis (Figure 3A). The priority of key proteins was analyzed by the degree of nodes output by the STRING database. Among them, the ALB value (degree=43) was the highest, followed by GAPDH (degree=39), CASP3 (degree=28), MAPK1 (degree=28), FN1 (degree=27), and IL-10 (degree=27), indicating that these proteins might act as bridges to other nodes in the PPI network (Figure 3B).
GENE ONTOLOGY FUNCTIONAL ENRICHMENT ANALYSIS:
To verify whether the 81 shared targets are associated with renal IR, GO functional enrichment analysis was carried out with R software 3.60, and the associated biological processes were clarified using the “DOSE” and “clusterProfiler” packages. Figure 4A displays the first 30 most significantly enriched GO terms, including biological processes (BP), molecular function (MF), and cellular component (CC) (P value ≤0.01). Supplementary Table 2 provides the P values, q-values, and counts of the remarkable enrichment items in BP, CC, and MF. The results indicate that the development of renal IR is regulated by many specific biological activities, including those involved in adenosine receptor signaling pathway (GO: 0001973), regulation of inflammatory response (GO: 0050727), regulation of body fluid levels (GO: 0050878), and response to hypoxia (GO: 0001666). Figure 4B presents the first 8 most significantly enriched GO terms in a circle-plot style. Among them, combined with the previous research results of our team [37], we focused on the adenosine receptor signaling pathway (GO: 0001973) and present the possible regulatory mechanism in Figure 5.
KEGG ENRICHMENT ANALYSIS OF PATHWAYS:
The pathways correlated to Cordyceps cicadae were obtained using the KEGG pathway enrichment analysis via R software 3.6.0. The significant pathways were enriched, and their corresponding P values were computed (calibrated by the Bonferroni method, the pathways with P values ≤0.01 were regarded as remarkable enrichment items). After sorting the P values, the first 30 pathways were shown in Figure 6A. Supplementary Table 3 provides the P values, q-values, and counts of the prominent enrichment pathways. The results indicated that multiple pathways participated in the process of renal IR, such as the cAMP signaling pathway (hsa04024), MAPK signaling pathway (hsa04010), PPAR signaling pathway (hsa03320), HIF-1 signaling pathway (hsa04066), and Rap1 signaling pathway (hsa04015). Figure 6B presents the first 8 most significantly enriched KEGG pathways in a circle-plot style.
Discussion
Renal IRI often occurs after recovery from renal ischemia, usually in septicemia, hypovolemic shock, and kidney transplantation [2]. In kidney transplant patients, IRI can lead to delayed graft function and cause a variety of long-term and short-term complications, thereby shortening the long-term survival rate of patients and bringing a heavy financial and medical burden to families and society [6,38]. Clinically, anti-oxidation, anti-inflammatory, inhibiting apoptosis, and improving renal perfusion are usually used for treatment. However, there is currently no treatment plan to completely avoid renal reperfusion injury. In China,
Cordycepin, adenosine, and cordycepic acid are the most important potential components of
With the results of I-T network analysis, 5 genes (TGM2, ERBB4, PTGIS, IGF1R, and SIRT3) were noted to be linked to the 3 active compounds identified, which indicated that these 3 components of
PPI network analysis showed that the protein node ALB had the highest value (degree=43), followed by GAPDH (degree=39), CASP3 (degree=28), MAPK1 (degree=28), FN1 (degree=27), and IL-10 (degree=27), indicating that these proteins might act as bridges connecting to other nodes in the PPI network (Figure 3B). In a related study, NO produced by IRI induced GAPDHS-nitrosylation at cysteine 150, which binds GAPDH to Siah1 (an E3 ubiquitinase), binds, promoting nuclear transformation of GAPDH, and then causing apoptosis [70]. Another study demonstrated that pre-silencing of GAPDH in cardiomyocyte IR by siRNA increased the autophagy in injured cells and the level of oxidative protective factors such as superoxide dismutase (SOD) and glutathione, and decreased the production of ROS, thereby alleviating the damage of cells and tissue caused by IRI [71]. CASP3 is a key protease involved in the caspase-dependent apoptosis pathway. It has been found that the expression of caspase-3 in renal IR increases significantly [72]. Deletion of the CASP3 gene can increase the levels of urinary cystatin C and serum creatinine and the increase the score of renal tubular injury in the early stage of AKI, and promote the death of renal tubular epithelial cells, while in long-term studies, it decreased the expression of α-smooth muscle actin and collagen deposition in peri-tubular capillaries, thus attenuating the degree of renal fibrosis [73]. Animal studies of renal fibrosis found that the expression of fibronectin 1 (FN1) generally increases with the increase of renal fibrosis [74]. Mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinases 1 and 2 (ERK1/2), the c-Jun N-terminal kinases (JNK), and p38 mitogen-activated protein kinases (p38-MAPK), participate in proliferation and differentiation in the process of renal growth and development, and also exert an important role in a variety of renal injuries such as inflammation, apoptosis, and fibrosis [75]. During IRI, ERK2 (MAPK1) can minimize H2O2-induced cell damage [76] and contribute to the healing of damaged renal tubular epithelial cells [77]. However, other scholars have found that decreasing the expression of ERK2 in mouse kidneys clearly diminished IRI and the production of associated inflammatory factors during renal IRI [78]. Interleukin (IL)-10 is a classic anti-inflammatory cytokine that can inhibit the production of various inflammation-related factors such as IL-1, TNF-α, IL-6, and IL-8 [79–82]. Meanwhile, in an animal study, the mRNA expression of IL-1, IL-6, TNF-α, and IL-18 was increased in mouse models of IL-10 gene knockout in the early stage of AKI [83]. Another study found that during the repair process of renal IRI, the expression of IL-10 increased, while the deletion of IL-10 gene increased the expression of pro-inflammatory factors such as TNF-α and IL-6, resulting in aggravated renal tissue injury [84]. In addition, increased expression of IL-10 has been confirmed to be effective in treatment of renal IRI [85,86].
The results of GO classification indicated that
According to the results of KEGG analysis, MAPK (hsa04010), cAMP (hsa04024), PPAR (hsa03320), Rap1 (hsa04015), and HIF-1 (hsa04066) signaling pathways are the primary pathways of
There are some deficiencies in the current research. First, the bioactive components actually absorbed by patients with renal IR may differ from those identified. Second, suppressor target genes and activated genes are difficult to distinguish. Furthermore, not all predictions were experimentally verified. Therefore, further research is needed to validate this TCM therapy.
Conclusions
The strategy of network pharmacology gives a new prediction method to mine the evidence of TCMs treatment mechanisms from an integrative point of view. Through target fishing and I-T network analysis, 3 active components of
Figures
 Figure 1. The 3D chemical structure models of the 3 components of cordycepin, adenosine and cordycepic acid. Figures obtained from PubChem (PubChem: https://pubchem.ncbi.nlm.nih.gov/), US National Institutes of Health.
Figure 1. The 3D chemical structure models of the 3 components of cordycepin, adenosine and cordycepic acid. Figures obtained from PubChem (PubChem: https://pubchem.ncbi.nlm.nih.gov/), US National Institutes of Health. 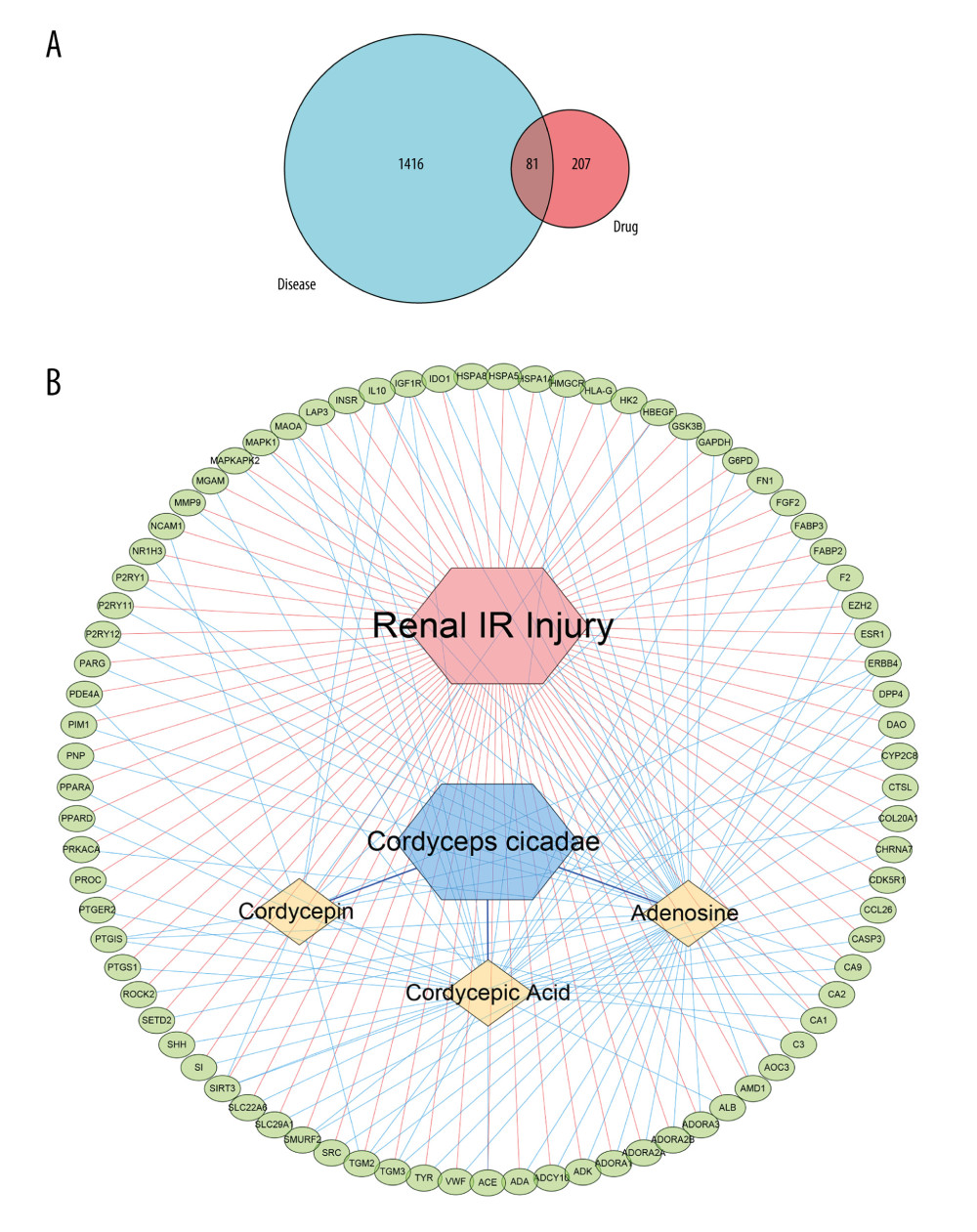 Figure 2. (A) The blue circle represents the related targets of renal ischemia/reperfusion injury (IRI), the red circle represents the related targets of Cordyceps cicadae, and the intersecting part of the 2 circles is the potential targets of Cordyceps cicadae acting on renal IRI. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (B) The ingredient–target (I-T) network in this study. The green nodes represent potential targets associated with renal IRI, the blue nodes represent Cordyceps cicadae, and the yellow nodes represent active ingredients of Cordyceps cicadae; the edges indicate the interactions among the nodes. Figure created by Cytoscape 3.7.2 software. IR – ischemia/reperfusion.
Figure 2. (A) The blue circle represents the related targets of renal ischemia/reperfusion injury (IRI), the red circle represents the related targets of Cordyceps cicadae, and the intersecting part of the 2 circles is the potential targets of Cordyceps cicadae acting on renal IRI. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (B) The ingredient–target (I-T) network in this study. The green nodes represent potential targets associated with renal IRI, the blue nodes represent Cordyceps cicadae, and the yellow nodes represent active ingredients of Cordyceps cicadae; the edges indicate the interactions among the nodes. Figure created by Cytoscape 3.7.2 software. IR – ischemia/reperfusion. 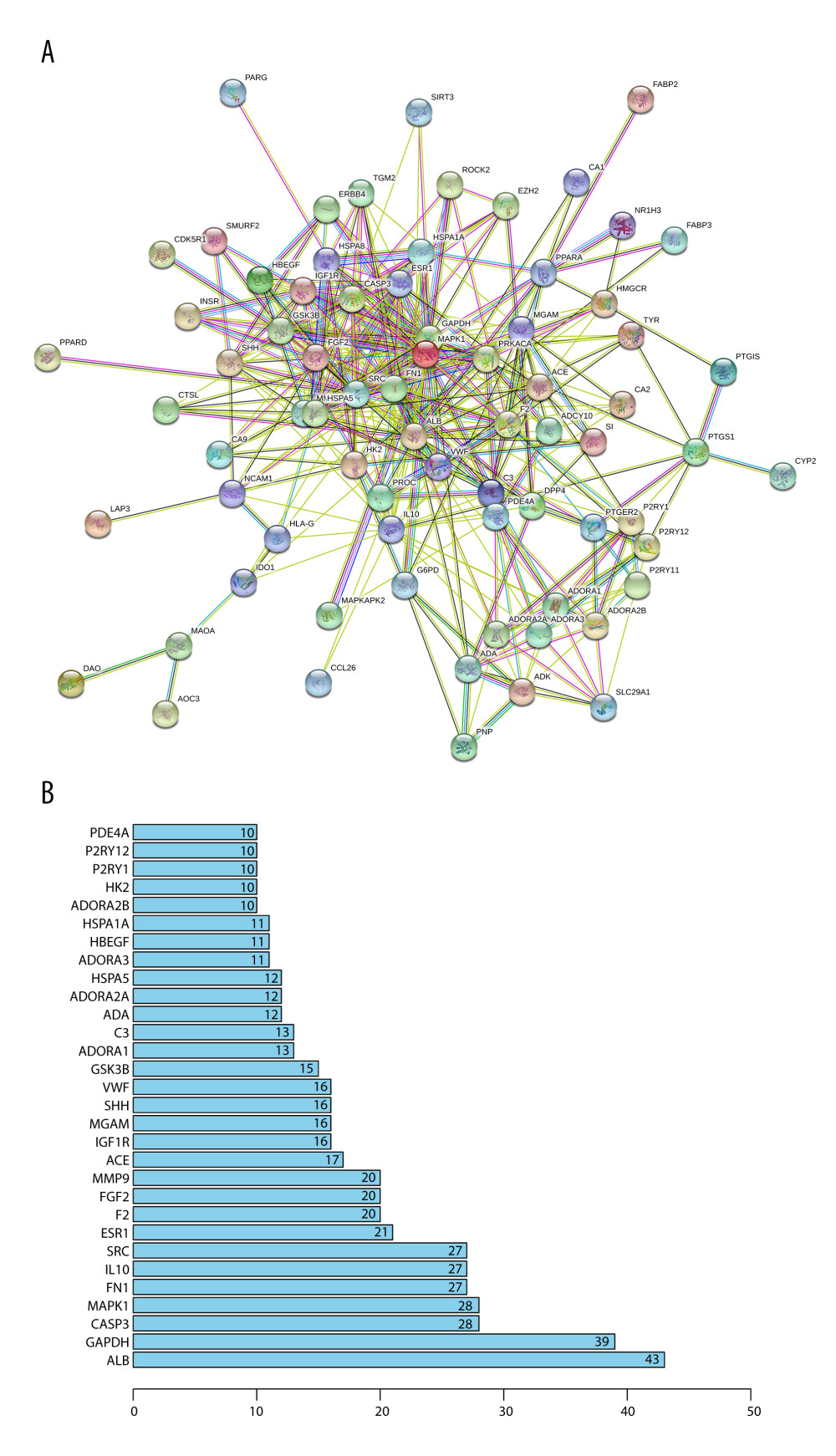 Figure 3. (A) The protein–protein interaction (PPI) network of potential targets correlated to Cordyceps cicadae against renal IRI. The nodes refer to proteins, and the edges indicate the interactions among them. The more edges on the same node, the greater the degree of the node. Figure created by STRING 11.0, https://string-db.org/. (B) Bar graph of the top 30 target proteins sorted by degree value. The longer the bar, the more significant the connection of the protein. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org).
Figure 3. (A) The protein–protein interaction (PPI) network of potential targets correlated to Cordyceps cicadae against renal IRI. The nodes refer to proteins, and the edges indicate the interactions among them. The more edges on the same node, the greater the degree of the node. Figure created by STRING 11.0, https://string-db.org/. (B) Bar graph of the top 30 target proteins sorted by degree value. The longer the bar, the more significant the connection of the protein. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). 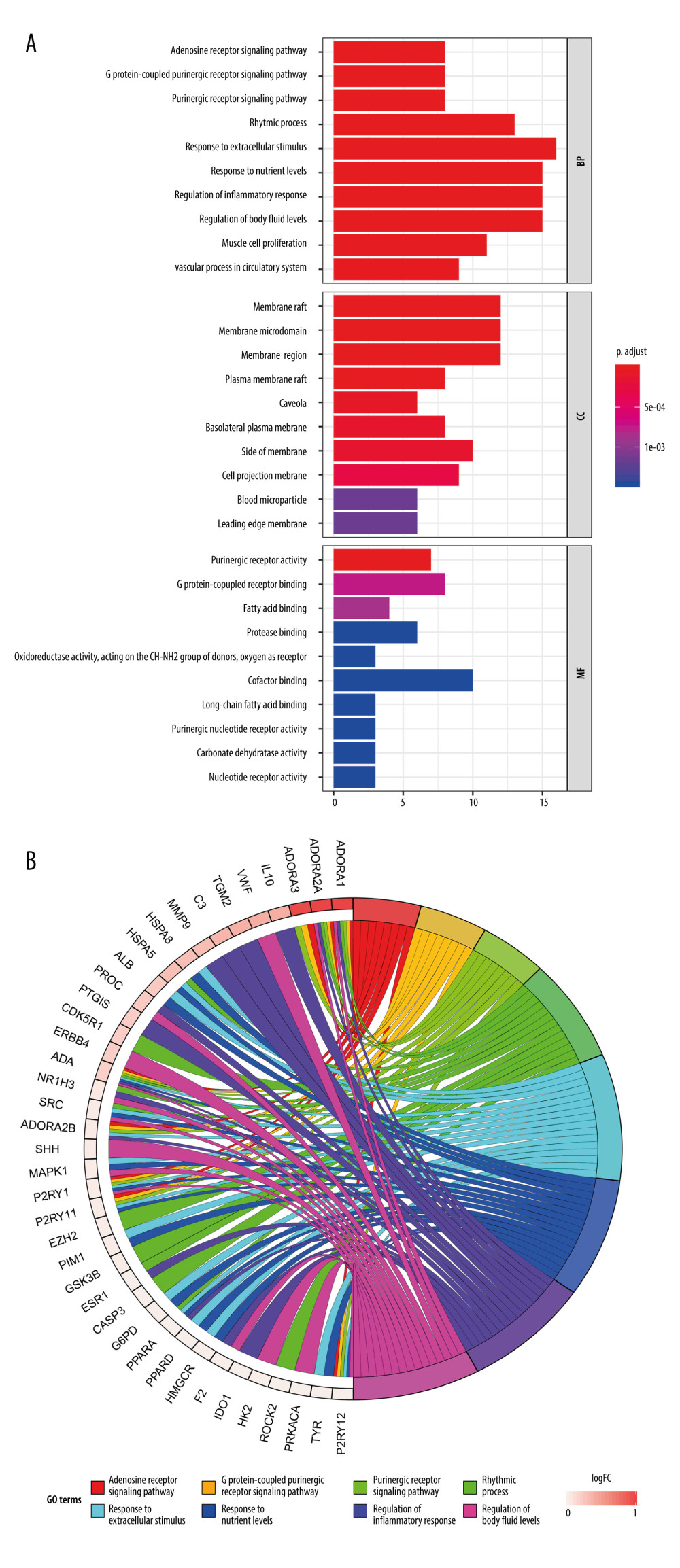 Figure 4. Gene Ontology (GO) analysis and significantly enriched GO terms of target genes related to Cordyceps cicadae in renal IR. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 GO terms in biological processes (BP), molecular function (MF) and cell component (CC). The x-axis represents significant enrichment counts of these terms, and the y-axis represents the categories of GO terms of the target genes (P value ≤0.01). (B) The first 8 significantly enriched GO terms in a circle-plot style. FC - Fold Change.
Figure 4. Gene Ontology (GO) analysis and significantly enriched GO terms of target genes related to Cordyceps cicadae in renal IR. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 GO terms in biological processes (BP), molecular function (MF) and cell component (CC). The x-axis represents significant enrichment counts of these terms, and the y-axis represents the categories of GO terms of the target genes (P value ≤0.01). (B) The first 8 significantly enriched GO terms in a circle-plot style. FC - Fold Change. 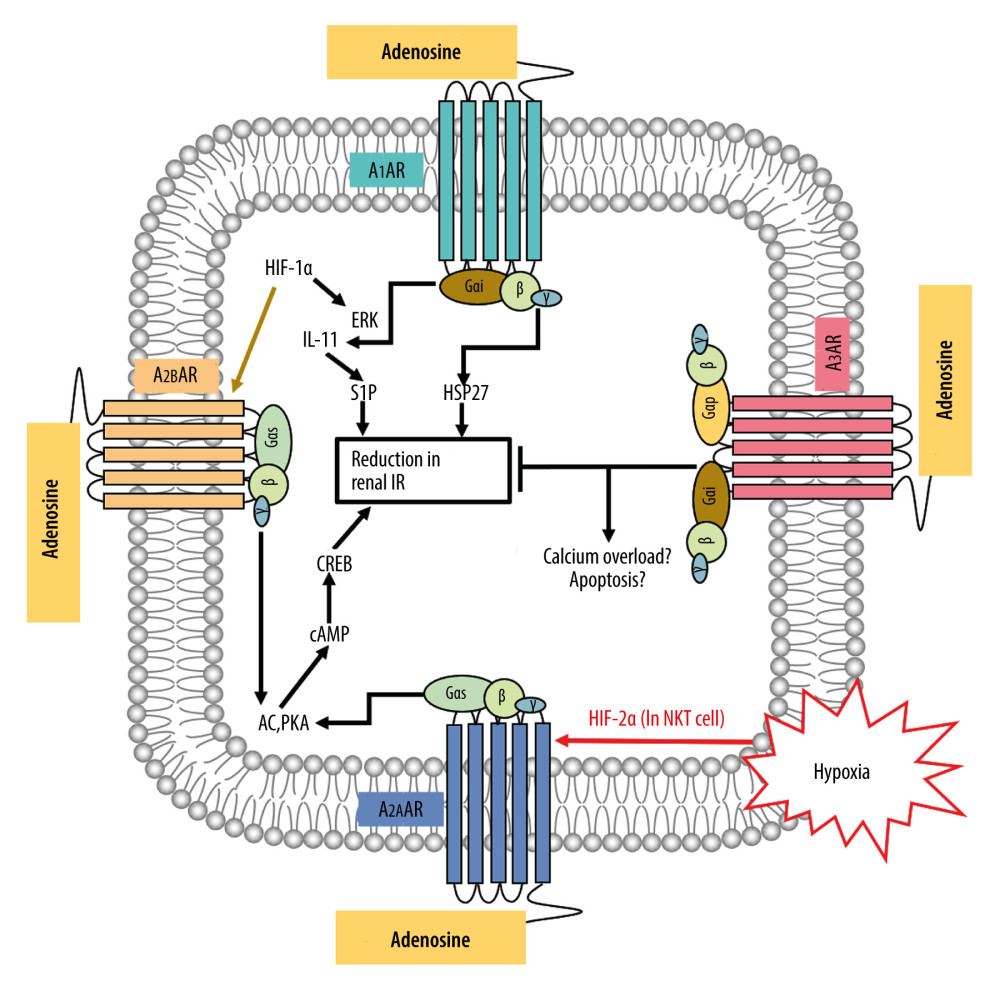 Figure 5. The possible regulatory mechanism of AR-mediated renal IRI. Figure created by PowerPoint for Microsoft Office 2016, Microsoft. AR – adenosine receptor; ERK – extracellular signal-regulated kinases; P38-MAPK – p38 mitogen-activated protein kinases; HIF-1α – hypoxia inducible factor 1 alpha; HIF-2α – hypoxia inducible factor 2 alpha; IL-11 – interleukin 11; S1P – sphingosine-1-phosphate; HSP27 – heat shock protein 27; AC – adenylate cyclase; PKA – protein kinase A; cAMP – adenosine 3′,5′ cyclic monophosphate; CREB – cAMP response-element binding protein; IR – ischemia/reperfusion.
Figure 5. The possible regulatory mechanism of AR-mediated renal IRI. Figure created by PowerPoint for Microsoft Office 2016, Microsoft. AR – adenosine receptor; ERK – extracellular signal-regulated kinases; P38-MAPK – p38 mitogen-activated protein kinases; HIF-1α – hypoxia inducible factor 1 alpha; HIF-2α – hypoxia inducible factor 2 alpha; IL-11 – interleukin 11; S1P – sphingosine-1-phosphate; HSP27 – heat shock protein 27; AC – adenylate cyclase; PKA – protein kinase A; cAMP – adenosine 3′,5′ cyclic monophosphate; CREB – cAMP response-element binding protein; IR – ischemia/reperfusion. 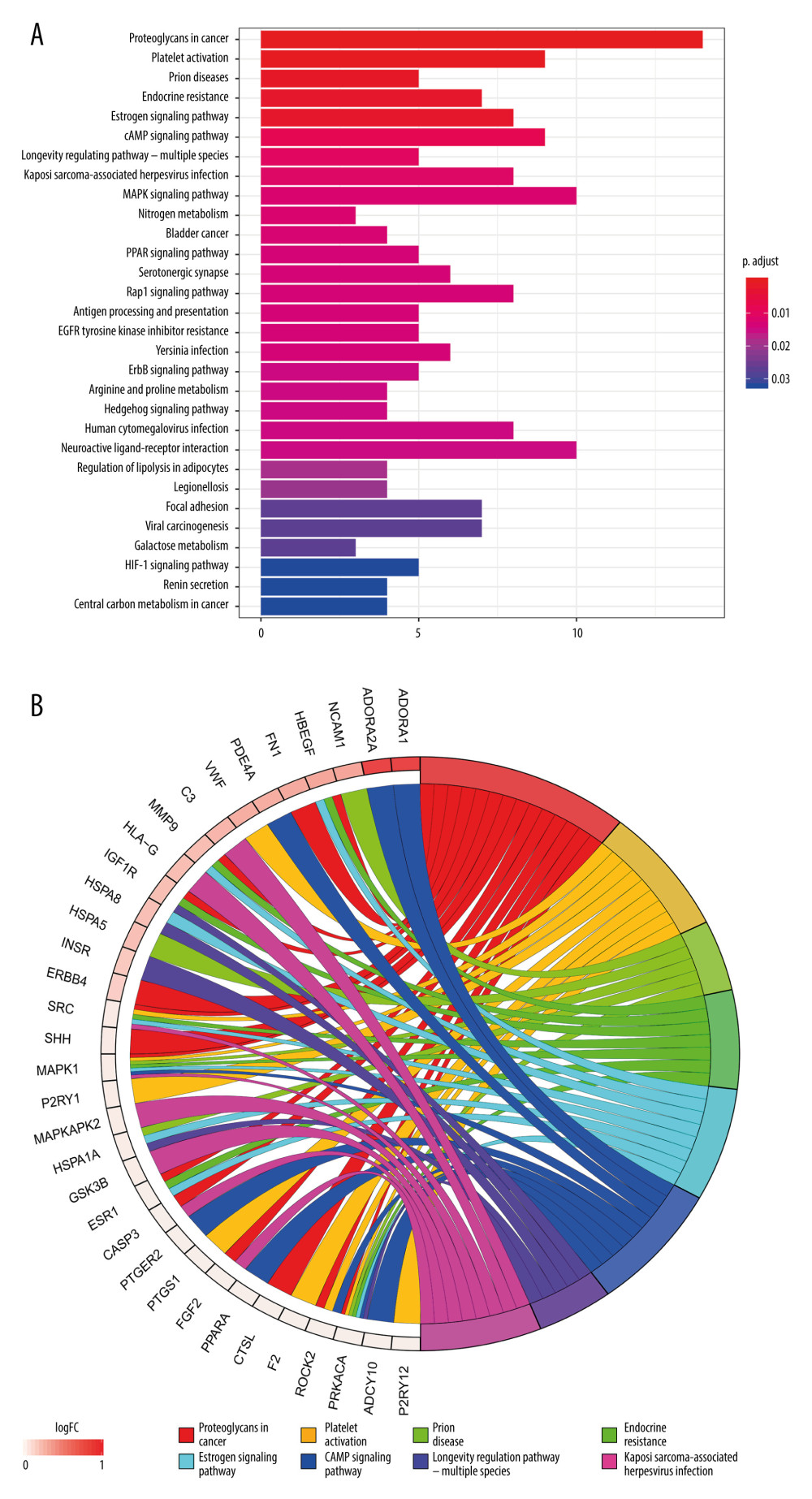 Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes related to renal IR that are modulated by Cordyceps cicadae. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 pathways sorted by P values. The x-axis represents significant enrichment counts of these pathways, and the y-axis represents the categories of KEGG pathways of the target genes (P value ≤0.01). (B) The top 8 significantly enriched KEGG pathways in a circle-plot style. FC – Fold Change.
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes related to renal IR that are modulated by Cordyceps cicadae. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 pathways sorted by P values. The x-axis represents significant enrichment counts of these pathways, and the y-axis represents the categories of KEGG pathways of the target genes (P value ≤0.01). (B) The top 8 significantly enriched KEGG pathways in a circle-plot style. FC – Fold Change. Tables
Supplementary Table 1. Information of Cordyceps cicadae and Renal IR-related targets. By combining the related targets of active ingredients of Cordyceps cicadae and the disease related targets, 81 overlapping targets were selected as the key targets in the treatment of renal IRI. Supplementary Table 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of therapeutic target genes and their corresponding counts, P value, and q-value. Through KEGG enrichment of the key targets, the remarkably (P value ≤0.01) enriched pathways were obtained, indicating that numerous targets were involved in the occurrence and development of renal IRI.
Supplementary Table 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of therapeutic target genes and their corresponding counts, P value, and q-value. Through KEGG enrichment of the key targets, the remarkably (P value ≤0.01) enriched pathways were obtained, indicating that numerous targets were involved in the occurrence and development of renal IRI.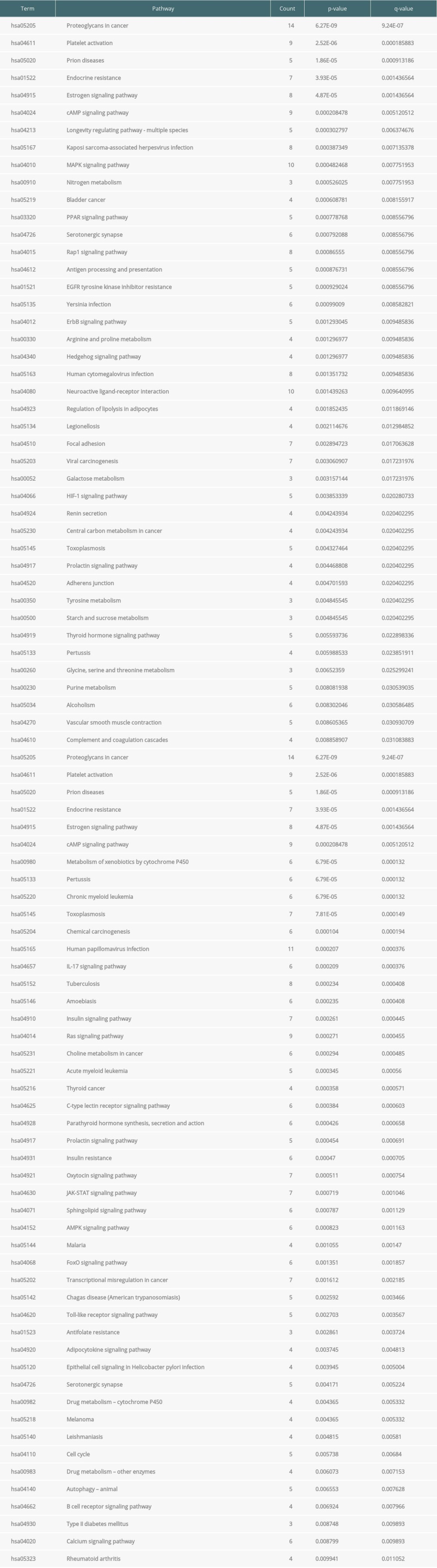
References
1. O’Kane D, Baldwin GS, Bolton DM, Preconditioning against renal ischaemia reperfusion injury: The failure to translate to the clinic: J Nephrol, 2019; 32(4); 539-47
2. Liu D, Tang S, Gan L, Cui W, Renal-protective effects and potential mechanisms of traditional chinese medicine after ischemia-reperfusion injury: Evid Based Complement Alternat Med, 2021; 2021; 5579327
3. Salvadori M, Rosso G, Bertoni E, Update on ischemia-reperfusion injury in kidney transplantation: Pathogenesis and treatment: World J Transplant, 2015; 5(2); 52-67
4. Wang L, Liu X, Chen H, Effect of picroside II on apoptosis induced by renal ischemia/reperfusion injury in rats: Exp Ther Med, 2015; 9(3); 817-22
5. Zhang J, Zou YR, Zhong X, Erythropoietin pretreatment ameliorates renal ischaemia-reperfusion injury by activating PI3K/Akt signalling: Nephrology (Carlton), 2015; 20(4); 266-72
6. Granata S, Votrico V, Spadaccino F, Oxidative stress and ischemia/reperfusion injury in kidney transplantation: Focus on ferroptosis, mitophagy and new antioxidants: Antioxidants (Basel), 2022; 11(4); 769
7. Bonavia A, Singbartl K, A review of the role of immune cells in acute kidney injury: Pediatr Nephrol, 2018; 33(10); 1629-39
8. Zheng L, Gao W, Hu C, Immune cells in ischemic acute kidney injury: Curr Protein Pept Sci, 2019; 20(8); 770-76
9. Sethi K, Rao K, Bolton D, Targeting HIF-1α to prevent renal ischemia-reperfusion injury: Does it work?: Int J Cell Biol, 2018; 2018; 9852791
10. Menting TP, Wever KE, Ozdemir-van Brunschot DM, Ischaemic preconditioning for the reduction of renal ischaemia reperfusion injury: Cochrane Database Syst Rev, 2017; 3(3); CD010777
11. Shang Z, Jiang Y, Guan X, Therapeutic effects of stem cells from different source on renal ischemia-reperfusion injury: A systematic review and network meta-analysis of animal studies: Front Pharmacol, 2021; 12; 713059
12. Girolami I, Gambaro G, Ghimenton C, Pre-implantation kidney biopsy: value of the expertise in determining histological score and comparison with the whole organ on a series of discarded kidneys: J Nephrol, 2020; 33(1); 167-76
13. Girolami I, Pantanowitz L, Marletta S, Artificial intelligence applications for pre-implantation kidney biopsy pathology practice: A systematic review: J Nephrol, 2022 [Online ahead of print]
14. Hermsen M, Volk V, Bräsen JH, Quantitative assessment of inflammatory infiltrates in kidney transplant biopsies using multiplex tyramide signal amplification and deep learning: Lab Invest, 2021; 101(8); 970-82
15. Palomino J, Echavarria R, Franco-Acevedo A, Opioids preconditioning upon renal function and ischemia-reperfusion injury: A narrative review: Medicina (Kaunas), 2019; 55(9); 522
16. Han SJ, Lee HT, Mechanisms and therapeutic targets of ischemic acute kidney injury: Kidney Res Clin Pract, 2019; 38(4); 427-40
17. Chen Z, Huang G-D, Recent developments in the experimental study of Chinese medicine for the treatment of renal ischemia-reperfusion injury: Guangxi Journal of Traditional Chinese Medicine, 2015; 38(05); 8-11
18. Zeng WB, Yu H, Ge F: Molecules, 2014; 19(5); 6123-41
19. Chu ZB, Chang J, Zhu Y, Sun X: Nat Prod Commun, 2015; 10(12); 2145-46
20. Lu MY, Chen CC, Lee LY: J Nat Prod, 2015; 78(10); 2452-60
21. Zheng R, Zhu R, Li X: Front Physiol, 2018; 9; 1229
22. Zhu R, Chen YP, Deng YY: J Zhejiang Univ Sci B, 2011; 12(12); 1024-33
23. Zhang J, Wen C, Duan Y: Int J Biol Macromol, 2019; 132; 906-14
24. Huang C, Zheng C, Li Y, Systems pharmacology in drug discovery and therapeutic insight for herbal medicines: Brief Bioinform, 2014; 15(5); 710-33
25. Berger SI, Iyengar R, Network analyses in systems pharmacology: Bioinformatics, 2009; 25(19); 2466-72
26. , UniProt: A worldwide hub of protein knowledge: Nucleic Acids Res, 2019; 47(D1); D506-15
27. Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D, GeneCards: Integrating information about genes, proteins and diseases: Trends Genet, 1997; 13(4); 163
28. Safran M, Dalah I, Alexander J, GeneCards Version 3: The human gene integrator: Database (Oxford), 2010; 2010; baq020
29. Shannon P, Markiel A, Ozier O, Cytoscape: A software environment for integrated models of biomolecular interaction networks: Genome Res, 2003; 13(11); 2498-504
30. Szklarczyk D, Gable AL, Lyon D, STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets: Nucleic Acids Res, 2019; 47(D1); D607-13
31. , Expansion of the Gene Ontology knowledgebase and resources: Nucleic Acids Res, 2017; 45(D1); D331-38
32. Kanehisa M, Goto S, KEGG: Kyoto encyclopedia of genes and genomes: Nucleic Acids Res, 2000; 28(1); 27-30
33. Kanehisa M, Furumichi M, Tanabe M, KEGG: New perspectives on genomes, pathways, diseases and drugs: Nucleic Acids Res, 2017; 45(D1); D353-61
34. Yu G, Wang LG, Yan GR, He QY, DOSE: An R/Bioconductor package for disease ontology semantic and enrichment analysis: Bioinformatics, 2015; 31(4); 608-9
35. Yu G, Wang LG, Han Y, He QY, clusterProfiler: An R package for comparing biological themes among gene clusters: Omics, 2012; 16(5); 284-87
36. Luo W, Brouwer C, Pathview: An R/Bioconductor package for pathway-based data integration and visualization: Bioinformatics, 2013; 29(14); 1830-31
37. Zhang J, Han C, Dai H, Hypoxia-inducible factor-2α limits natural killer T cell cytotoxicity in renal ischemia/reperfusion injury: J Am Soc Nephrol, 2016; 27(1); 92-106
38. Zhao H, Alam A, Soo AP, Ischemia-reperfusion injury reduces long term renal graft survival: Mechanism and beyond: EBioMedicine, 2018; 28; 31-42
39. Liu Y-N, Chen Y-P, Wang L-H, The effect of cordyceps sobolifera mycelium on renal tubulointerstitial expression of PAI-1 and UPA in rat with unilateral ureter obstruction: Chinese Journal of Integrated Traditional and Western Nephrology, 2012; 13(03); 197-200
40. Deng JS, Jiang WP, Chen CC: Oxid Med Cell Longev, 2020; 2020; 7912763
41. Huang YS, Wang X, Feng Z: Evid Based Complement Alternat Med, 2020; 2020; 7202519
42. Hsu JH, Jhou BY, Yeh SH: J Nutr Food Sci, 2015; 5; 432
43. Li L, Zhang T, Li C: Drug Des Devel Ther, 2019; 13; 103-17
44. Yap SC, Lee HT, Adenosine and protection from acute kidney injury: Curr Opin Nephrol Hypertens, 2012; 21(1); 24-32
45. Pandey S, Aggarwal D, Gupta K, “Adenosine an old player with new possibilities in kidney diseases”: Preclinical evidences and clinical perspectives: Life Sci, 2021; 265; 118834
46. Cao W, Wan H, Wu L, Adenosine kinase inhibition attenuates ischemia reperfusion-induced acute kidney injury: Life Sci, 2020; 256; 117972
47. Do T, Nguyen HC: BioTechnologia, 2019; 100(3); 219-26
48. Lugo-Baruqui JA, Ayyathurai R, Use of mannitol for ischemia reperfusion injury in kidney transplant and partial nephrectomies-review of literature: Curr Urol Rep, 2019; 20(1); 6
49. Aydin HR, Sekerci CA, Yigit E, Protective effect of cordycepin on experimental renal ischemia/reperfusion injury in rats: Arch Ital Urol Androl, 2020; 92(4); 2020.4.340
50. Han F, Dou M, Wang Y, Cordycepin protects renal ischemia/reperfusion injury through regulating inflammation, apoptosis, and oxidative stress: Acta Biochim Biophys Sin (Shanghai), 2020; 52(2); 125-32
51. Tuli HS, Sharma AK, Sandhu SS, Kashyap D, Cordycepin: A bioactive metabolite with therapeutic potential: Life Sci, 2013; 93(23); 863-69
52. Furini G, Burhan I, Huang L, Insights into the heparan sulphate-dependent externalisation of transglutaminase-2 (TG2) in glucose-stimulated proximal-like tubular epithelial cells: Anal Biochem, 2020; 603; 113628
53. Grenard P, Bresson-Hadni S, El Alaoui S, Transglutaminase-mediated cross-linking is involved in the stabilization of extracellular matrix in human liver fibrosis: J Hepatol, 2001; 35(3); 367-75
54. Wang Z, Xu W, Kang J, Overexpression of alfalfa Orange gene in tobacco enhances carotenoid accumulation and tolerance to multiple abiotic stresses: Plant Physiol Biochem, 2018; 130; 613-22
55. El Nahas AM, Abo-Zenah H, Skill NJ, Elevated epsilon-(gamma-glutamyl)lysine in human diabetic nephropathy results from increased expression and cellular release of tissue transglutaminase: Nephron Clin Pract, 2004; 97(3); c108-17
56. Ohlmeier S, Nieminen P, Gao J, Lung tissue proteomics identifies elevated transglutaminase 2 levels in stable chronic obstructive pulmonary disease: Am J Physiol Lung Cell Mol Physiol, 2016; 310(11); L1155-65
57. Zakrzewicz A, Atanasova S, Padberg W, Grau V, Monocytic tissue transglutaminase in a rat model for reversible acute rejection and chronic renal allograft injury: Mediators Inflamm, 2015; 2015; 429653
58. Iwamoto R, Mekada E, ErbB and HB-EGF signaling in heart development and function: Cell Struct Funct, 2006; 31(1); 1-14
59. Zeng F, Miyazawa T, Kloepfer LA, Harris RC, ErbB4 deletion accelerates renal fibrosis following renal injury: Am J Physiol Renal Physiol, 2018; 314(5); F773-87
60. Klein T, Klaus G, Kömhoff M, Prostacyclin synthase: Upregulation during renal development and in glomerular disease as well as its constitutive expression in cultured human mesangial cells: Mediators Inflamm, 2015; 2015; 654151
61. Cao Y, Guan Y, Xu YY, Hao CM, Endothelial prostacyclin protects the kidney from ischemia-reperfusion injury: Pflugers Arch, 2019; 471(4); 543-55
62. Pollak M, Insulin-like growth factor physiology and cancer risk: Eur J Cancer, 2000; 36(10); 1224-28
63. Chisalita SI, Arnqvist HJ, Insulin-like growth factor I receptors are more abundant than insulin receptors in human micro- and macrovascular endothelial cells: Am J Physiol Endocrinol Metab, 2004; 286(6); E896-901
64. Liang M, Woodard LE, Liang A, Protective role of insulin-like growth factor-1 receptor in endothelial cells against unilateral ureteral obstruction-induced renal fibrosis: Am J Pathol, 2015; 185(5); 1234-50
65. Morigi M, Perico L, Benigni A, Sirtuins in renal health and disease: J Am Soc Nephrol, 2018; 29(7); 1799-809
66. Perico L, Morigi M, Benigni A, Mitochondrial sirtuin 3 and renal diseases: Nephron, 2016; 134(1); 14-19
67. Kim D, Park W, Lee S, Absence of Sirt3 aggravates cisplatin nephrotoxicity via enhanced renal tubular apoptosis and inflammation: Mol Med Rep, 2018; 18(4); 3665-72
68. Wang Q, Xu J, Li X, Sirt3 modulate renal ischemia-reperfusion injury through enhancing mitochondrial fusion and activating the ERK-OPA1 signaling pathway: J Cell Physiol, 2019; 234(12); 23495-506
69. Ouyang J, Zeng Z, Fang H, SIRT3 inactivation promotes acute kidney injury through elevated acetylation of SOD2 and p53: J Surg Res, 2019; 233; 221-30
70. Li C, Feng JJ, Wu YP, Zhang GY, Cerebral ischemia-reperfusion induces GAPDH S-nitrosylation and nuclear translocation: Biochemistry (Mosc), 2012; 77(6); 671-78
71. Liang S, Aiqun M, Figtree G, Ping Z, GAPDH-silence preserves H9C2 cells from acute hypoxia and reoxygenation injury: Int J Biol Macromol, 2015; 81; 375-86
72. Yang B, Jain S, Ashra SY, Apoptosis and caspase-3 in long-term renal ischemia/reperfusion injury in rats and divergent effects of immunosuppressants: Transplantation, 2006; 81(10); 1442-50
73. Yang B, Lan S, Dieudé M, Caspase-3 is a pivotal regulator of microvascular rarefaction and renal fibrosis after ischemia-reperfusion injury: J Am Soc Nephrol, 2018; 29(7); 1900-16
74. Stribos EGD, Luangmonkong T, Leliveld AM, Precision-cut human kidney slices as a model to elucidate the process of renal fibrosis: Transl Res, 2016; 170; 8-16e1
75. Ma FY, Sachchithananthan M, Flanc RS, Nikolic-Paterson DJ, Mitogen activated protein kinases in renal fibrosis: Front Biosci (Schol Ed), 2009; 1(1); 171-87
76. Park KM, Fogelgren B, Zuo X, Exocyst Sec10 protects epithelial barrier integrity and enhances recovery following oxidative stress, by activation of the MAPK pathway: Am J Physiol Renal Physiol, 2010; 298(3); F818-26
77. Jang HS, Han SJ, Kim JI, Activation of ERK accelerates repair of renal tubular epithelial cells, whereas it inhibits progression of fibrosis following ischemia/reperfusion injury: Biochim Biophys Acta, 2013; 1832(12); 1998-2008
78. Ka SO, Hwang HP, Jang JH, The protein kinase 2 inhibitor tetrabromobenzotriazole protects against renal ischemia reperfusion injury: Sci Rep, 2015; 5; 14816
79. Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A, Interleukin-10 and the interleukin-10 receptor: Annu Rev Immunol, 2001; 19; 683-765
80. Scholz J, Lukacs-Kornek V, Engel DR, Renal dendritic cells stimulate IL-10 production and attenuate nephrotoxic nephritis: J Am Soc Nephrol, 2008; 19(3); 527-37
81. Takeshita S, Gage JR, Kishimoto T, Differential regulation of IL-6 gene transcription and expression by IL-4 and IL-10 in human monocytic cell lines: J Immunol, 1996; 156(7); 2591-98
82. de Waal Malefyt R, Abrams J, Bennett B, Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: An autoregulatory role of IL-10 produced by monocytes: J Exp Med, 1991; 174(5); 1209-20
83. Sakai K, Nozaki Y, Murao Y, Protective effect and mechanism of IL-10 on renal ischemia-reperfusion injury: Lab Invest, 2019; 99(5); 671-83
84. Wan X, Huang WJ, Chen W, IL-10 deficiency increases renal ischemia-reperfusion injury: Nephron Exp Nephrol, 2014; 128(1–2); 37-45
85. Tang TT, Wang B, Wu M, Extracellular vesicle-encapsulated IL-10 as novel nanotherapeutics against ischemic AKI: Sci Adv, 2020; 6(33); eaaz0748
86. Patil CN, Wallace K, LaMarca BD, Low-dose testosterone protects against renal ischemia-reperfusion injury by increasing renal IL-10-to-TNF-α ratio and attenuating T-cell infiltration: Am J Physiol Renal Physiol, 2016; 311(2); F395-403
87. Malek M, Nematbakhsh M, Renal ischemia/reperfusion injury; from pathophysiology to treatment: J Renal Inj Prev, 2015; 4(2); 20-27
88. Jang HR, Ko GJ, Wasowska BA, Rabb H, The interaction between ischemia-reperfusion and immune responses in the kidney: J Mol Med (Berl), 2009; 87(9); 859-64
89. Stroo I, Stokman G, Teske GJ, Chemokine expression in renal ischemia/reperfusion injury is most profound during the reparative phase: Int Immunol, 2010; 22(6); 433-42
90. Chen Y, Yang SH, Hueng DY, Cordycepin induces apoptosis of C6 glioma cells through the adenosine 2A receptor-p53-caspase-7-PARP pathway: Chem Biol Interact, 2014; 216; 17-25
91. Leu SF, Poon SL, Pao HY, Huang BM, The in vivo and in vitro stimulatory effects of cordycepin on mouse leydig cell steroidogenesis: Biosci Biotechnol Biochem, 2011; 75(4); 723-31
92. Day YJ, Huang L, McDuffie MJ, Renal protection from ischemia mediated by A2A adenosine receptors on bone marrow-derived cells: J Clin Invest, 2003; 112(6); 883-91
93. Day YJ, Li Y, Rieger JM, Ramos SI, A2A adenosine receptors on bone marrow-derived cells protect liver from ischemia–zreperfusion injury: J Immunol, 2005; 174(8); 5040-46
94. Grenz A, Osswald H, Eckle T, The reno-vascular A2B adenosine receptor protects the kidney from ischemia: PLoS Med, 2008; 5(6); e137
95. Kong T, Westerman KA, Faigle M, HIF-dependent induction of adenosine A2B receptor in hypoxia: FASEB J, 2006; 20(13); 2242-50
96. Lee HT, Ota-Setlik A, Xu H, A3 adenosine receptor knockout mice are protected against ischemia- and myoglobinuria-induced renal failure: Am J Physiol Renal Physiol, 2003; 284(2); F267-73
97. Lee HT, Emala CW, Protective effects of renal ischemic preconditioning and adenosine pretreatment: Role of A(1) and A(3) receptors: Am J Physiol Renal Physiol, 2000; 278(3); F380-87
98. Stridh S, Palm F, Hansell P, Renal interstitial hyaluronan: Functional aspects during normal and pathological conditions: Am J Physiol Regul Integr Comp Physiol, 2012; 302(11); R1235-49
99. Wang XB, Liu P, Tang ZPCordyceps mycelia extract decreases portal hypertension in rats with dimethylnitrosamine-induced liver cirrhosis: A study on its histological basis: Zhong Xi Yi Jie He Xue Bao, 2008; 6(11); 1136-44 [in Chinese]
100. Wang M, Xu H, Chong Lee Shin OL, Compound α-keto acid tablet supplementation alleviates chronic kidney disease progression via inhibition of the NF-κB and MAPK pathways: J Transl Med, 2019; 17(1); 122
101. Chu W, Li M, Li F, Immediate splenectomy down-regulates the MAPK-NF-κB signaling pathway in rat brain after severe traumatic brain injury: J Trauma Acute Care Surg, 2013; 74(6); 1446-53
102. Zmonarski SC, Banasik M, Madziarska K, The role of toll-like receptors in multifactorial mechanisms of early and late renal allotransplant injury, with a focus on the TLR4 receptor and mononuclear cells: Adv Clin Exp Med, 2019; 28(7); 981-87
103. Fan Y, Meng S, Wang Y, Visfatin/PBEF/Nampt induces EMMPRIN and MMP-9 production in macrophages via the NAMPT-MAPK (p38, ERK1/2)-NF-κB signaling pathway: Int J Mol Med, 2011; 27(4); 607-15
104. Guo X, Jiang H, Chen J, RP105 ameliorates hypoxia/reoxygenation injury in cardiac microvascular endothelial cells by suppressing TLR4/MAPKs/NF-κB signaling: Int J Mol Med, 2018; 42(1); 505-13
105. Fawzy MA, Maher SA, Bakkar SM, Pantoprazole attenuates MAPK (ERK1/2, JNK, p38)-NF-κB and apoptosis signaling pathways after renal ischemia/reperfusion injury in rats: Int J Mol Sci, 2021; 22(19); 10669
106. Yu H, Zhou Q, Huang R, Yuan MEffect of Cordyceps sinensis on the expression of HIF-1α and NGAL in rats with renal ischemia-reperfusion injury: Zhong Nan Da Xue Xue Bao Yi Xue Ban, 2012; 37(1); 57-66 [in Chinese]
107. Rosenberger C, Heyman SN, Rosen S, Up-regulation of HIF in experimental acute renal failure: Evidence for a protective transcriptional response to hypoxia: Kidney Int, 2005; 67(2); 531-42
108. Sun X, Tan Y, Dong C, HIF-1α therapy enhances collateral vascularization in a ischemia model in hindlimb of rabbits: Chinese Journal of Laboratory Diagnosis, 2008; 12(12); 1504-6
109. Leonard MO, Cottell DC, Godson C, The role of HIF-1 alpha in transcriptional regulation of the proximal tubular epithelial cell response to hypoxia: J Biol Chem, 2003; 278(41); 40296-304
110. Skalhegg BS, Tasken K, Specificity in the cAMP/PKA signaling pathway. Differential expression,regulation, and subcellular localization of subunits of PKA: Front Biosci, 2000; 5; D678-93
111. Thapa K, Singh TG, Kaur A, Cyclic nucleotide phosphodiesterase inhibition as a potential therapeutic target in renal ischemia reperfusion injury: Life Sci, 2021; 282; 119843
112. Fung JC, Yue GG, Fung KP, Cordyceps militaris extract stimulates Cl(−) secretion across human bronchial epithelia by both Ca(2+)(−) and cAMP-dependent pathways: J Ethnopharmacol, 2011; 138(1); 201-11
113. Chen YC, Huang YL, Huang BM, Cordyceps sinensis mycelium activates PKA and PKC signal pathways to stimulate steroidogenesis in MA-10 mouse Leydig tumor cells: Int J Biochem Cell Biol, 2005; 37(1); 214-23
114. Tian X, Li Y, Shen Y, Apoptosis and inhibition of proliferation of cancer cells induced by cordycepin: Oncol Lett, 2015; 10(2); 595-99
115. Brown JD, Plutzky J, Peroxisome proliferator-activated receptors as transcriptional nodal points and therapeutic targets: Circulation, 2007; 115(4); 518-33
116. Hennuyer N, Tailleux A, Torpier G, PPARalpha, but not PPARgamma, activators decrease macrophage-laden atherosclerotic lesions in a nondiabetic mouse model of mixed dyslipidemia: Arterioscler Thromb Vasc Biol, 2005; 25(9); 1897-902
117. Li AC, Binder CJ, Gutierrez A, Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma: J Clin Invest, 2004; 114(11); 1564-76
118. Pane B, Gazzola V, Spinella G, Inflammatory response modulation through a PPARγ agonist during surgically induced visceral ischemia in an animal model: Ann Vasc Surg, 2018; 48; 189-94
119. He H, Tang J, Ru D, Protective effects of Cordyceps extract against UVB-induced damage and prediction of application prospects in the topical administration: An experimental validation and network pharmacology study: Biomed Pharmacother, 2020; 121; 109600
120. Takahashi S, Tamai M, Nakajima S, Blockade of adipocyte differentiation by cordycepin: Br J Pharmacol, 2012; 167(3); 561-75
121. Enserink JM, Christensen AE, de Rooij J, A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK: Nat Cell Biol, 2002; 4(11); 901-6
122. Shah S, Brock EJ, Ji K, Mattingly RR, Ras and Rap1: A tale of two GTPases: Semin Cancer Biol, 2019; 54; 29-39
123. El-Mokadem BM, El-Abhar HS, Abdallah DM, Epac-1/Rap-1 signaling pathway orchestrates the reno-therapeutic effect of ticagrelor against renal ischemia/reperfusion model: Biomed Pharmacother, 2021; 139; 111488
124. Chen YL, Yeh SH, Lin TW: Int J Med Mushrooms, 2015; 17(8); 771-81
125. Zhang HW, Lin ZX, Tung YS: Cochrane Database Syst Rev, 2014(12); CD008353
126. Chen B, Sun Y, Luo F, Wang C, Bioactive metabolites and potential mycotoxins produced by cordyceps fungi: A review of safety: Toxins (Basel), 2020; 12(6); 410
127. Bibi S, Wang YB, Tang DX, Prospects for discovering the secondary metabolites of cordyceps sensu lato by the integrated strategy: Med Chem, 2021; 17(2); 97-120
Figures
 Figure 1. The 3D chemical structure models of the 3 components of cordycepin, adenosine and cordycepic acid. Figures obtained from PubChem (PubChem: https://pubchem.ncbi.nlm.nih.gov/), US National Institutes of Health.
Figure 1. The 3D chemical structure models of the 3 components of cordycepin, adenosine and cordycepic acid. Figures obtained from PubChem (PubChem: https://pubchem.ncbi.nlm.nih.gov/), US National Institutes of Health. Figure 2. (A) The blue circle represents the related targets of renal ischemia/reperfusion injury (IRI), the red circle represents the related targets of Cordyceps cicadae, and the intersecting part of the 2 circles is the potential targets of Cordyceps cicadae acting on renal IRI. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (B) The ingredient–target (I-T) network in this study. The green nodes represent potential targets associated with renal IRI, the blue nodes represent Cordyceps cicadae, and the yellow nodes represent active ingredients of Cordyceps cicadae; the edges indicate the interactions among the nodes. Figure created by Cytoscape 3.7.2 software. IR – ischemia/reperfusion.
Figure 2. (A) The blue circle represents the related targets of renal ischemia/reperfusion injury (IRI), the red circle represents the related targets of Cordyceps cicadae, and the intersecting part of the 2 circles is the potential targets of Cordyceps cicadae acting on renal IRI. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (B) The ingredient–target (I-T) network in this study. The green nodes represent potential targets associated with renal IRI, the blue nodes represent Cordyceps cicadae, and the yellow nodes represent active ingredients of Cordyceps cicadae; the edges indicate the interactions among the nodes. Figure created by Cytoscape 3.7.2 software. IR – ischemia/reperfusion. Figure 3. (A) The protein–protein interaction (PPI) network of potential targets correlated to Cordyceps cicadae against renal IRI. The nodes refer to proteins, and the edges indicate the interactions among them. The more edges on the same node, the greater the degree of the node. Figure created by STRING 11.0, https://string-db.org/. (B) Bar graph of the top 30 target proteins sorted by degree value. The longer the bar, the more significant the connection of the protein. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org).
Figure 3. (A) The protein–protein interaction (PPI) network of potential targets correlated to Cordyceps cicadae against renal IRI. The nodes refer to proteins, and the edges indicate the interactions among them. The more edges on the same node, the greater the degree of the node. Figure created by STRING 11.0, https://string-db.org/. (B) Bar graph of the top 30 target proteins sorted by degree value. The longer the bar, the more significant the connection of the protein. Figure created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). Figure 4. Gene Ontology (GO) analysis and significantly enriched GO terms of target genes related to Cordyceps cicadae in renal IR. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 GO terms in biological processes (BP), molecular function (MF) and cell component (CC). The x-axis represents significant enrichment counts of these terms, and the y-axis represents the categories of GO terms of the target genes (P value ≤0.01). (B) The first 8 significantly enriched GO terms in a circle-plot style. FC - Fold Change.
Figure 4. Gene Ontology (GO) analysis and significantly enriched GO terms of target genes related to Cordyceps cicadae in renal IR. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 GO terms in biological processes (BP), molecular function (MF) and cell component (CC). The x-axis represents significant enrichment counts of these terms, and the y-axis represents the categories of GO terms of the target genes (P value ≤0.01). (B) The first 8 significantly enriched GO terms in a circle-plot style. FC - Fold Change. Figure 5. The possible regulatory mechanism of AR-mediated renal IRI. Figure created by PowerPoint for Microsoft Office 2016, Microsoft. AR – adenosine receptor; ERK – extracellular signal-regulated kinases; P38-MAPK – p38 mitogen-activated protein kinases; HIF-1α – hypoxia inducible factor 1 alpha; HIF-2α – hypoxia inducible factor 2 alpha; IL-11 – interleukin 11; S1P – sphingosine-1-phosphate; HSP27 – heat shock protein 27; AC – adenylate cyclase; PKA – protein kinase A; cAMP – adenosine 3′,5′ cyclic monophosphate; CREB – cAMP response-element binding protein; IR – ischemia/reperfusion.
Figure 5. The possible regulatory mechanism of AR-mediated renal IRI. Figure created by PowerPoint for Microsoft Office 2016, Microsoft. AR – adenosine receptor; ERK – extracellular signal-regulated kinases; P38-MAPK – p38 mitogen-activated protein kinases; HIF-1α – hypoxia inducible factor 1 alpha; HIF-2α – hypoxia inducible factor 2 alpha; IL-11 – interleukin 11; S1P – sphingosine-1-phosphate; HSP27 – heat shock protein 27; AC – adenylate cyclase; PKA – protein kinase A; cAMP – adenosine 3′,5′ cyclic monophosphate; CREB – cAMP response-element binding protein; IR – ischemia/reperfusion. Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes related to renal IR that are modulated by Cordyceps cicadae. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 pathways sorted by P values. The x-axis represents significant enrichment counts of these pathways, and the y-axis represents the categories of KEGG pathways of the target genes (P value ≤0.01). (B) The top 8 significantly enriched KEGG pathways in a circle-plot style. FC – Fold Change.
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the target genes related to renal IR that are modulated by Cordyceps cicadae. Figures created by R 3.6.0 with Rstudio, R: The R Project for Statistical Computing (r-project.org). (A) The top 30 pathways sorted by P values. The x-axis represents significant enrichment counts of these pathways, and the y-axis represents the categories of KEGG pathways of the target genes (P value ≤0.01). (B) The top 8 significantly enriched KEGG pathways in a circle-plot style. FC – Fold Change. Tables
 Supplementary Table 1. Information of Cordyceps cicadae and Renal IR-related targets. By combining the related targets of active ingredients of Cordyceps cicadae and the disease related targets, 81 overlapping targets were selected as the key targets in the treatment of renal IRI.
Supplementary Table 1. Information of Cordyceps cicadae and Renal IR-related targets. By combining the related targets of active ingredients of Cordyceps cicadae and the disease related targets, 81 overlapping targets were selected as the key targets in the treatment of renal IRI. Supplementary Table 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of therapeutic target genes and their corresponding counts, P value, and q-value. Through KEGG enrichment of the key targets, the remarkably (P value ≤0.01) enriched pathways were obtained, indicating that numerous targets were involved in the occurrence and development of renal IRI.
Supplementary Table 3. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of therapeutic target genes and their corresponding counts, P value, and q-value. Through KEGG enrichment of the key targets, the remarkably (P value ≤0.01) enriched pathways were obtained, indicating that numerous targets were involved in the occurrence and development of renal IRI. In Press
15 Mar 2024 : Review article
Approaches and Challenges in the Current Management of Cytomegalovirus in Transplant Recipients: Highlighti...Ann Transplant In Press; DOI: 10.12659/AOT.941185
18 Mar 2024 : Original article
Does Antibiotic Use Increase the Risk of Post-Transplantation Diabetes Mellitus? A Retrospective Study of R...Ann Transplant In Press; DOI: 10.12659/AOT.943282
20 Mar 2024 : Original article
Transplant Nephrectomy: A Comparative Study of Timing and Techniques in a Single InstitutionAnn Transplant In Press; DOI: 10.12659/AOT.942252
28 Mar 2024 : Original article
Association Between FEV₁ Decline Rate and Mortality in Long-Term Follow-Up of a 21-Patient Pilot Clinical T...Ann Transplant In Press; DOI: 10.12659/AOT.942823
Most Viewed Current Articles
05 Apr 2022 : Original article
Impact of Statins on Hepatocellular Carcinoma Recurrence After Living-Donor Liver TransplantationDOI :10.12659/AOT.935604
Ann Transplant 2022; 27:e935604
12 Jan 2022 : Original article
Risk Factors for Developing BK Virus-Associated Nephropathy: A Single-Center Retrospective Cohort Study of ...DOI :10.12659/AOT.934738
Ann Transplant 2022; 27:e934738
22 Nov 2022 : Original article
Long-Term Effects of Everolimus-Facilitated Tacrolimus Reduction in Living-Donor Liver Transplant Recipient...DOI :10.12659/AOT.937988
Ann Transplant 2022; 27:e937988
15 Mar 2022 : Case report
Combined Liver, Pancreas-Duodenum, and Kidney Transplantation for Patients with Hepatitis B Cirrhosis, Urem...DOI :10.12659/AOT.935860
Ann Transplant 2022; 27:e935860








